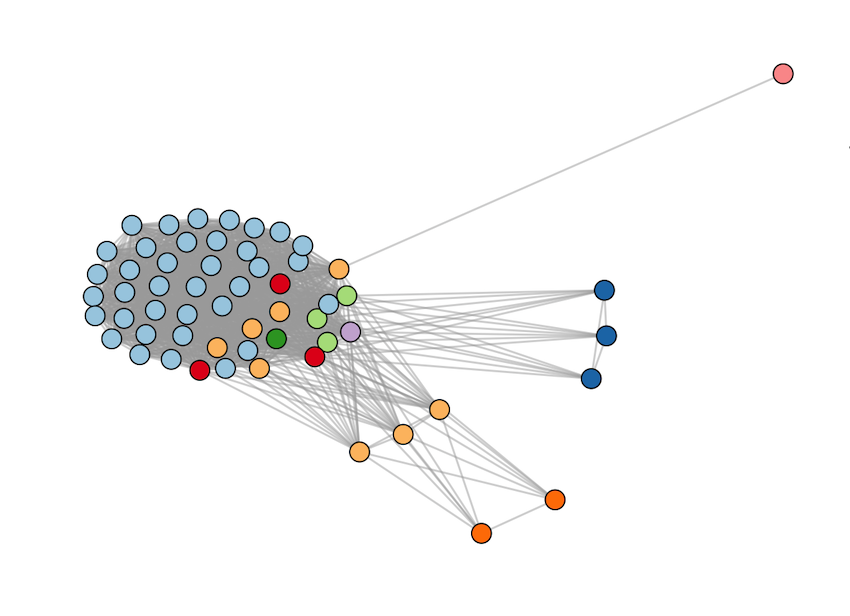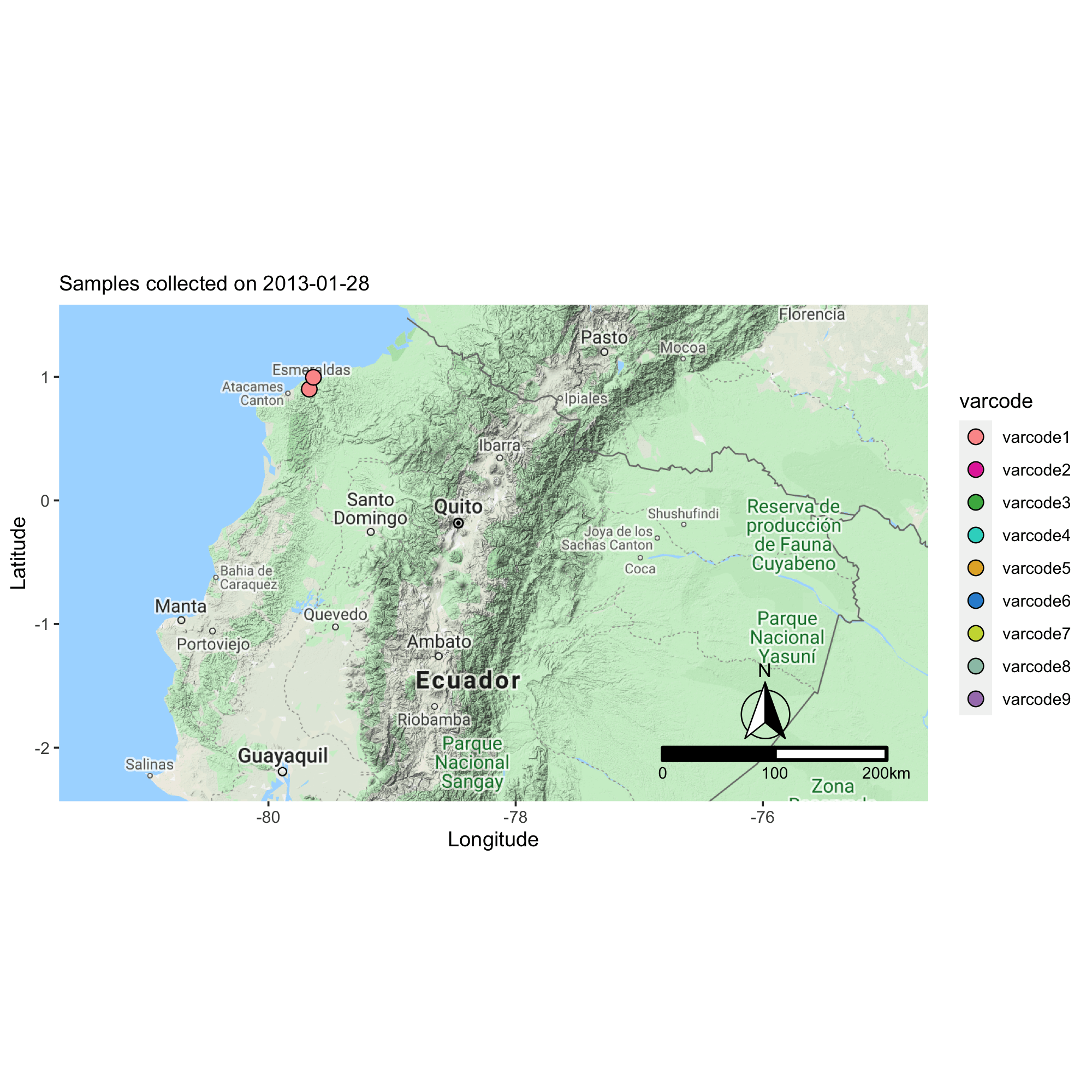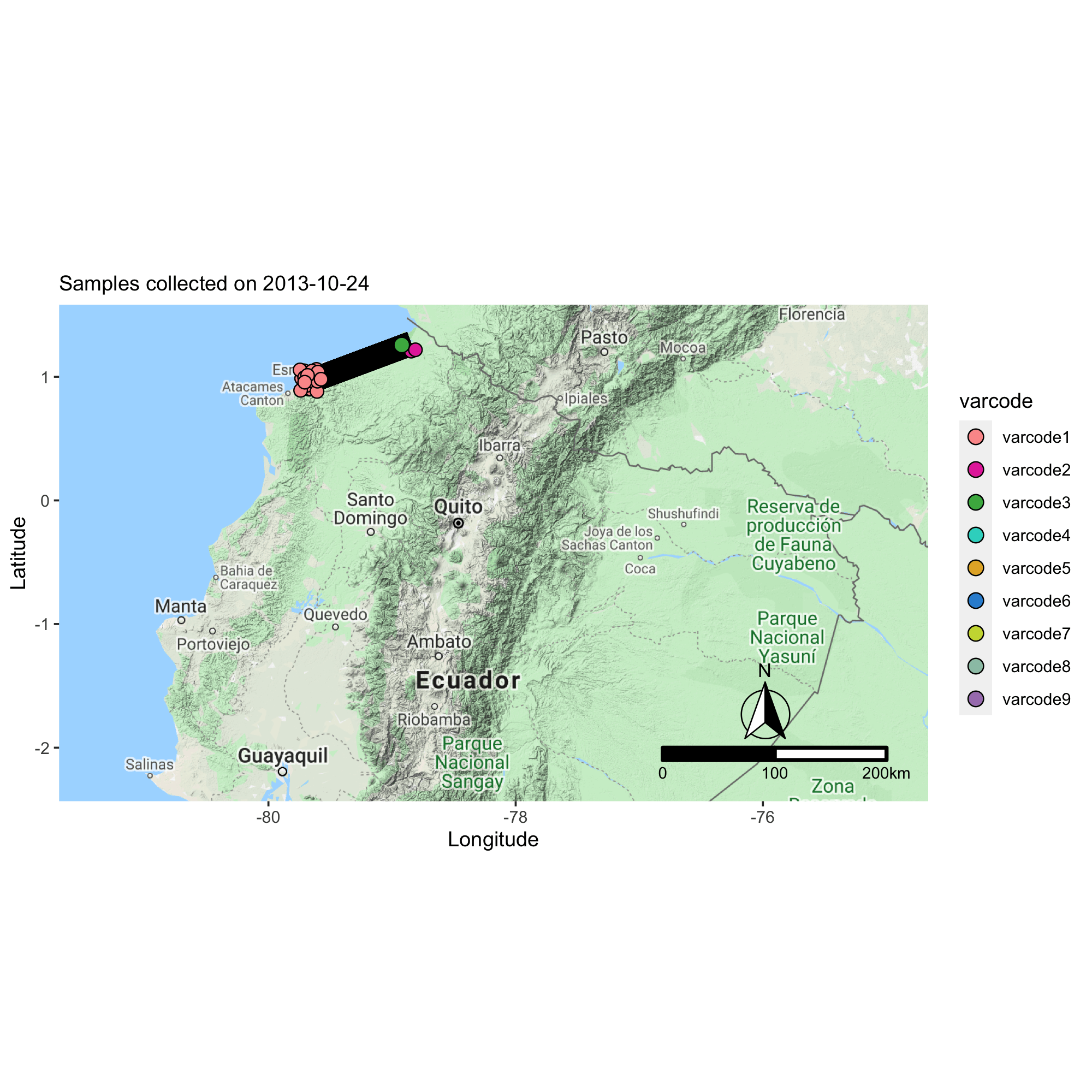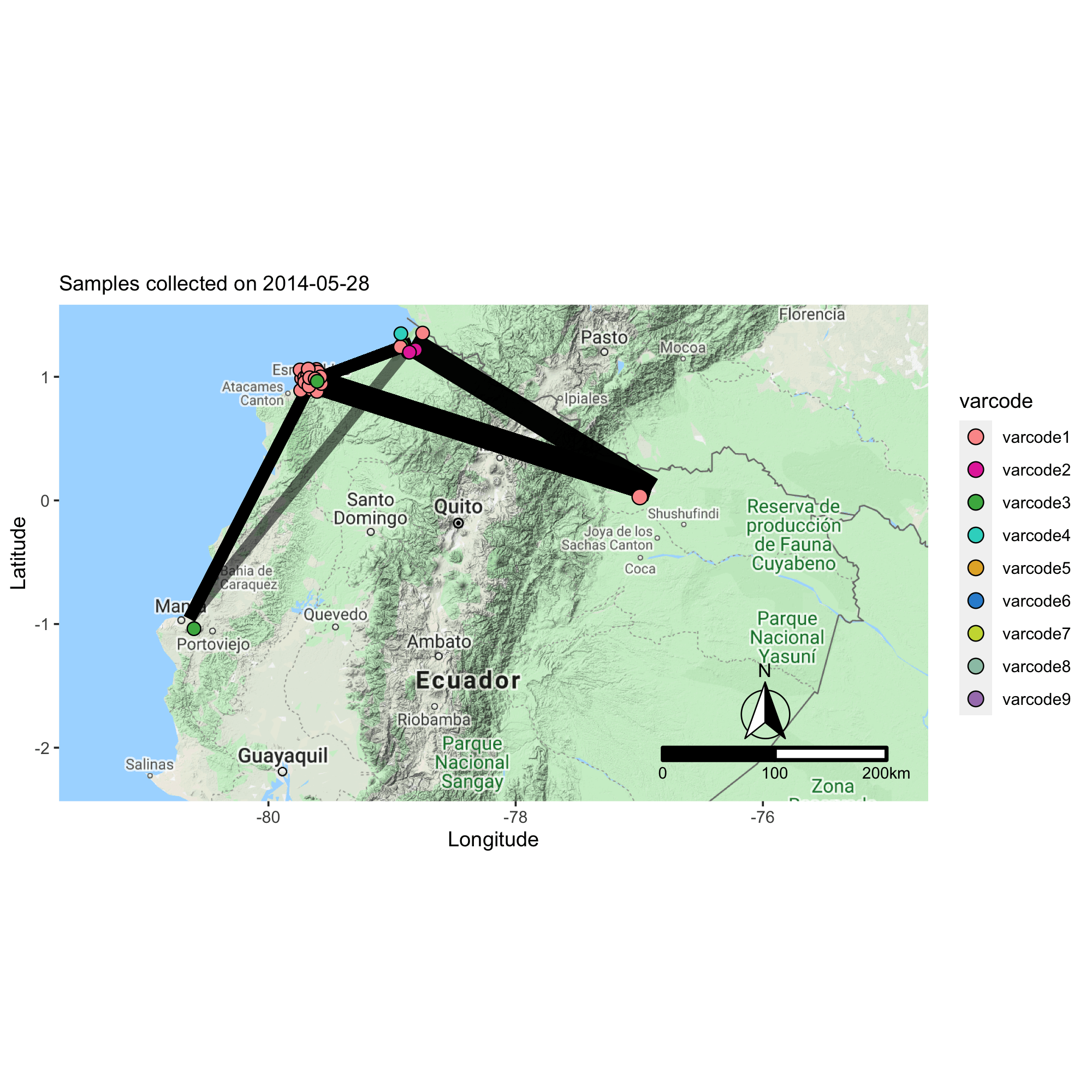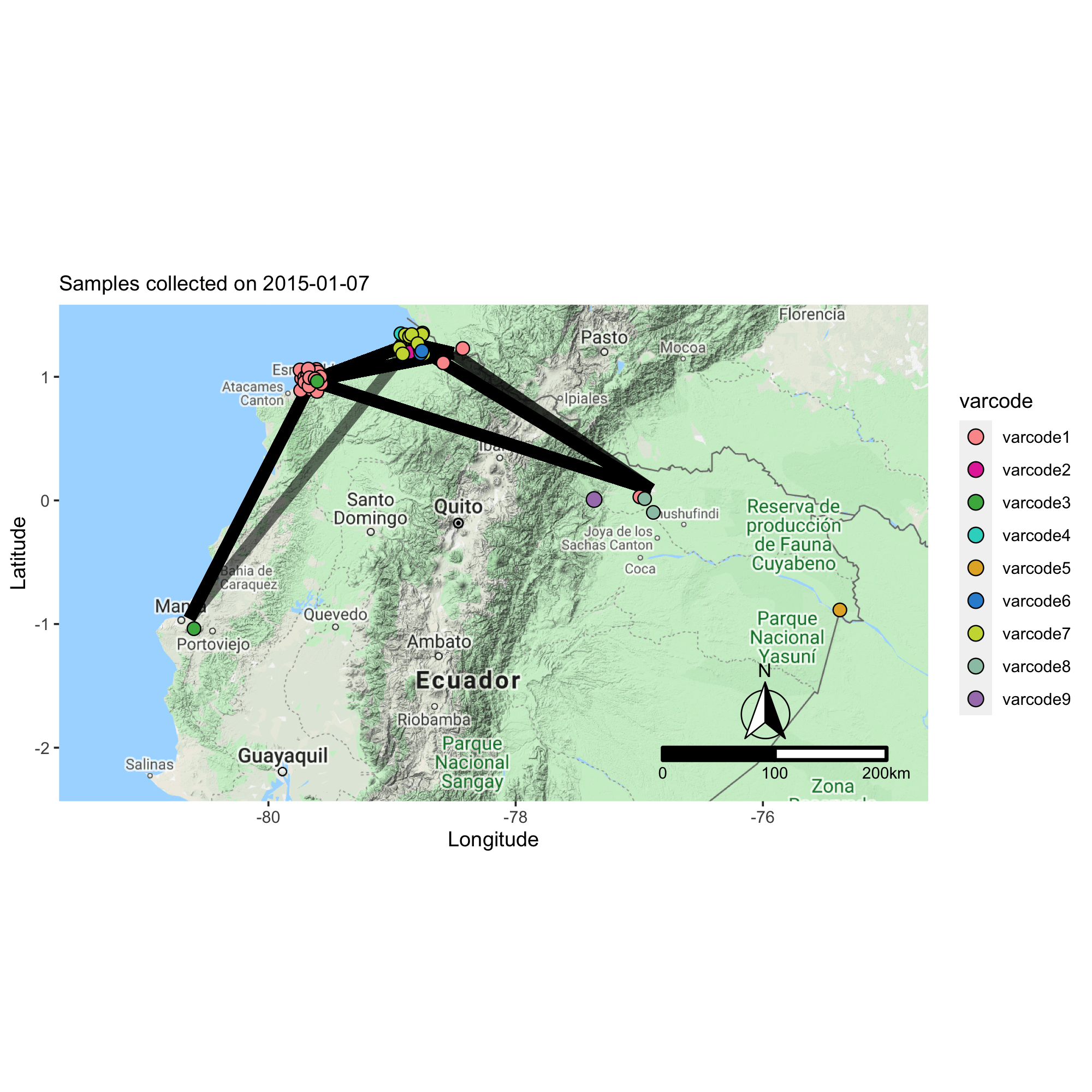
Applying the varcode for malaria outbreak surveillance in Ecuador
This collaborative project aims to describe the genomic epidemiology of continued disease transmission of P. falciparum in Ecuador after considerable progress towards elimination in previous years. By applying novel genetic tools, we aim to better understand factors underlying continued transmission after an outbreak in Ecuador by investigating parasite microevolution and transmission dynamics to inform whether malaria cases in Ecuador are locally-acquired or imported from neighboring countries.